About labs
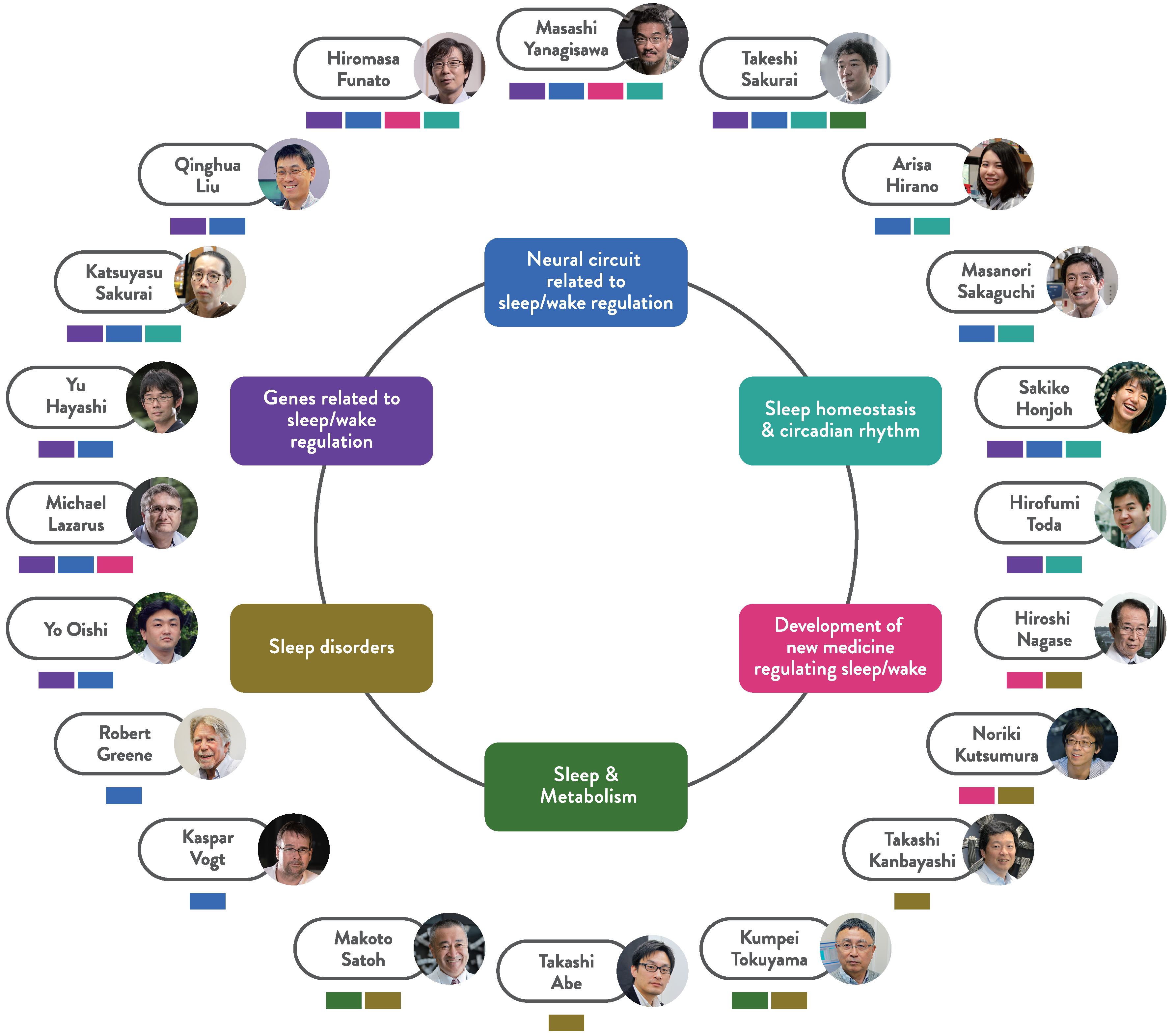
There are fourteen core labs and eight satellite labs in IIIS, covering various research fields e.g. molecular genetics, neuroscience, physiology, pharmaceutical science, human sleep physiology, etc. Those labs collaborate with each other to drive forward innovative sleep research.